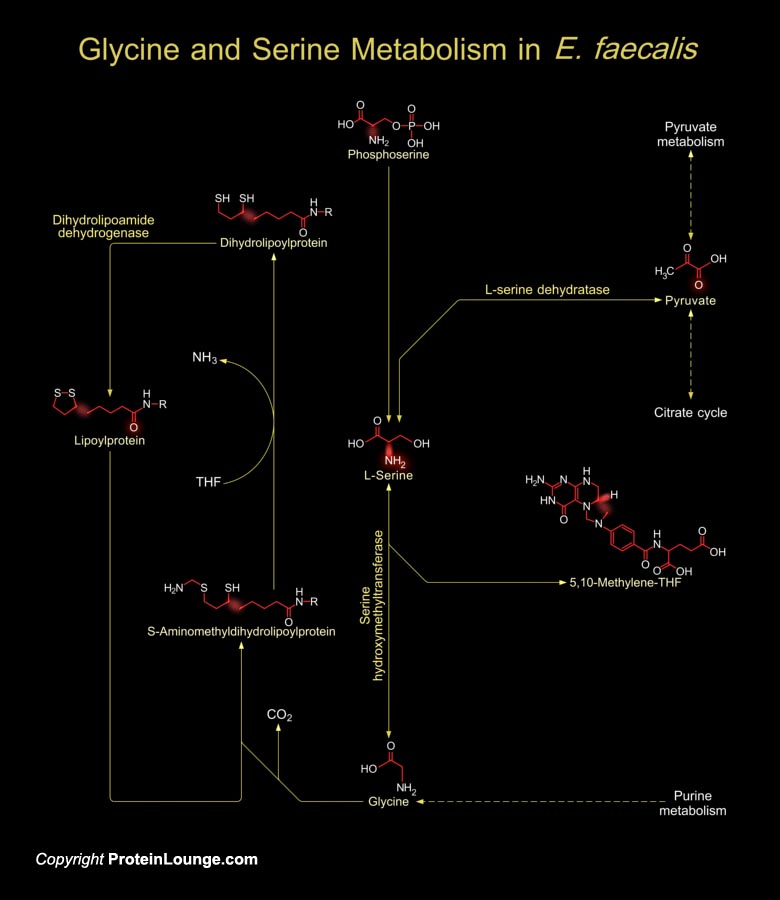
E. faecalis (Enterococcus faecalis), also known as S. faecalis (Streptococcus faecalis), a Gram-positive bacterium, is a natural inhabitant of the mammalian gastrointestinal tract and is found in soil, sewage, water and food, frequently through fecal contamination. It is an opportunistic pathogen that is a major cause of urinary tract infections, Bacteremia, bloodstream infections, wound infections, and infective Endocarditis. It has become a nosocomial pathogen that is refractory to most therapeutic options (Ref.1). The involvement of D- and L-Amino acid metabolism like L-Serine, L-Threonine and Glycine plays a major role in cell sustenance, generation of several essential compounds, intact protein synthesis, and pathogenesis in E. faecalis (Ref.2). In E.[..]
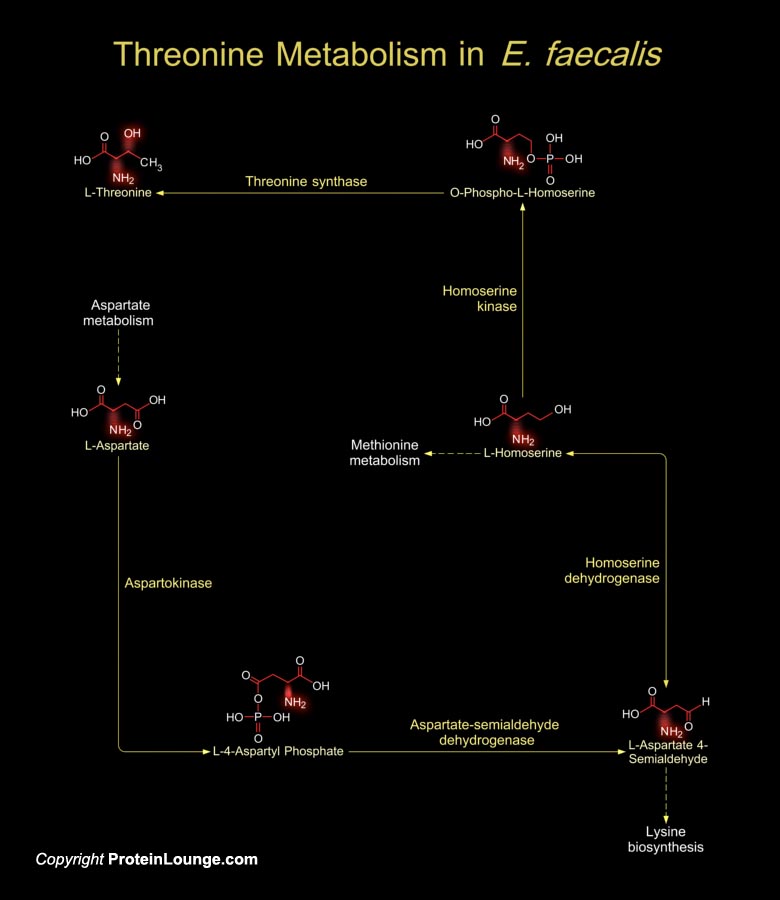
Enterococci are Gram-positive, facultative anaerobic and Lactic acid producing bacteria. Most strains are non-hemolytic. E. faecalis (Enterococcus faecalis), also known as S. faecalis (Streptococcus faecalis), the second most frequent enterococcal species, is a saprophytic commensal that inhabits the oral cavity and gastrointestinal flora of humans and animals and behaves as an opportunistic pathogen causing severe urinary tract infections, surgical wound infections, Bacteremia, and bacterial Endocarditis. The increased incidence of E. faecalis infection has been related to the innate resistance of this microorganism to many commonly used antimicrobial agents and to its ability to become resistant to most, and in some cases to all, of the presently available[..]
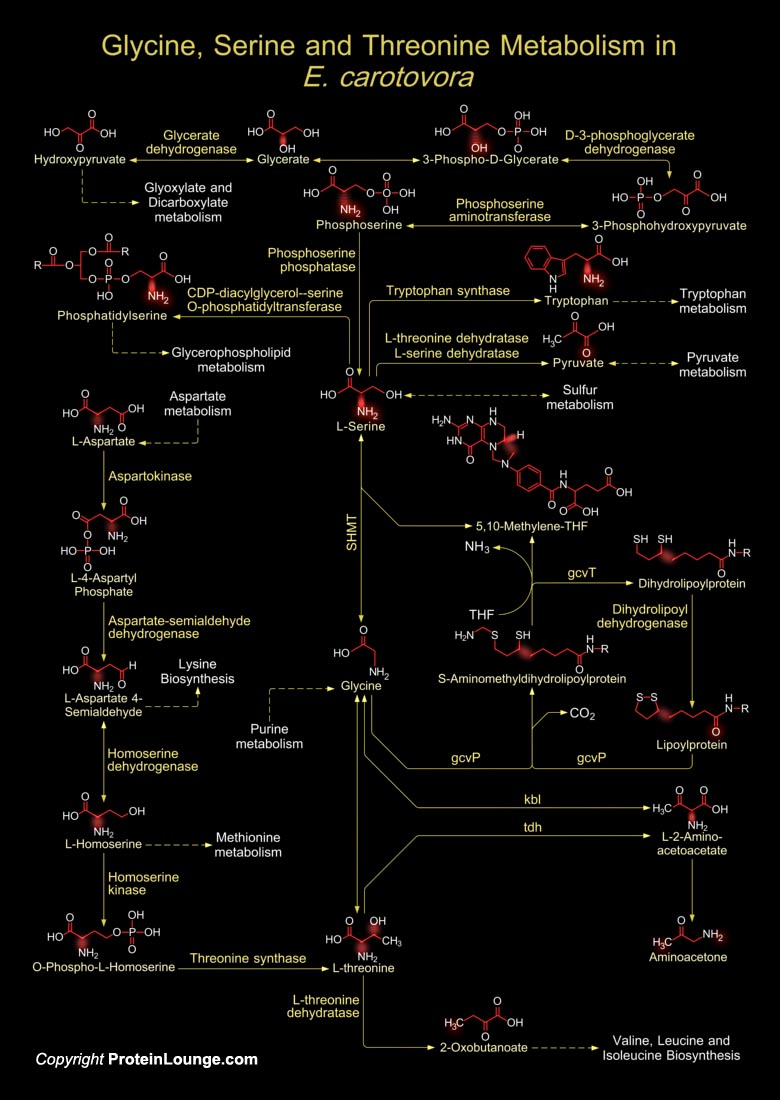
E. carotovora (Erwinia carotovora) is a species of plant pathogenic, Gram-negative, facultatively anaerobic, rod-shaped bacteria which gets its name from carrots, but it affects many other vegetables, including potatoes, cucumbers, onions, tomatoes, lettuce and even some ornamental plants like Iris. The bacterial family Enterobacteriaceae is notable for its well studied human pathogens, including Salmonella, Yersinia, Shigella, and Escherichia spp. However, it also contains several plant pathogens. Erwinia carotovora subsp. atroseptica (Eca) strain SCRI1043 is the causative agent of Soft rot and Blackleg potato diseases (Ref.1). Gram-negative bacteria employ a type of conserved signaling called Quorum Sensing. Quorum Sensing regulates as a switch controlling metabolic[..]
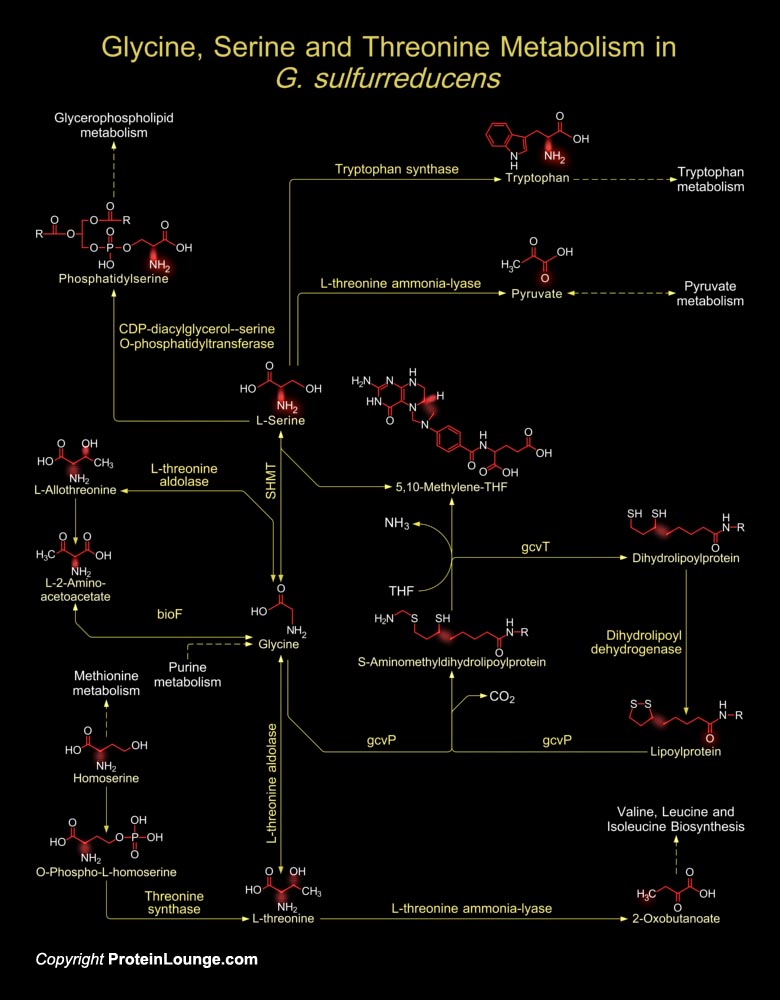
G. sulfurreducens (Geobacter sulfurreducens), a delta-proteobacterium, is an obligately anaerobic, non-fermentative, non-motile, Gram-negative rod. Geobacter species are of interest because of their novel electron transfer capabilities, impact on the natural environment and their application to the Bioremediation of contaminated environments and harvesting electricity from waste organic matter. G. sulfurreducens breaks down heavy metals and is being used to clean up toxic metal waste sites like Uranium, etc. Central to the metabolism of G. sulfurreducens is the ability to anaerobically oxidize Acetate (an abundant electron donor and carbon source in subsurface zones) completely to CO2 (Carbondioxide) and water using a variety of electron acceptors including metal[..]
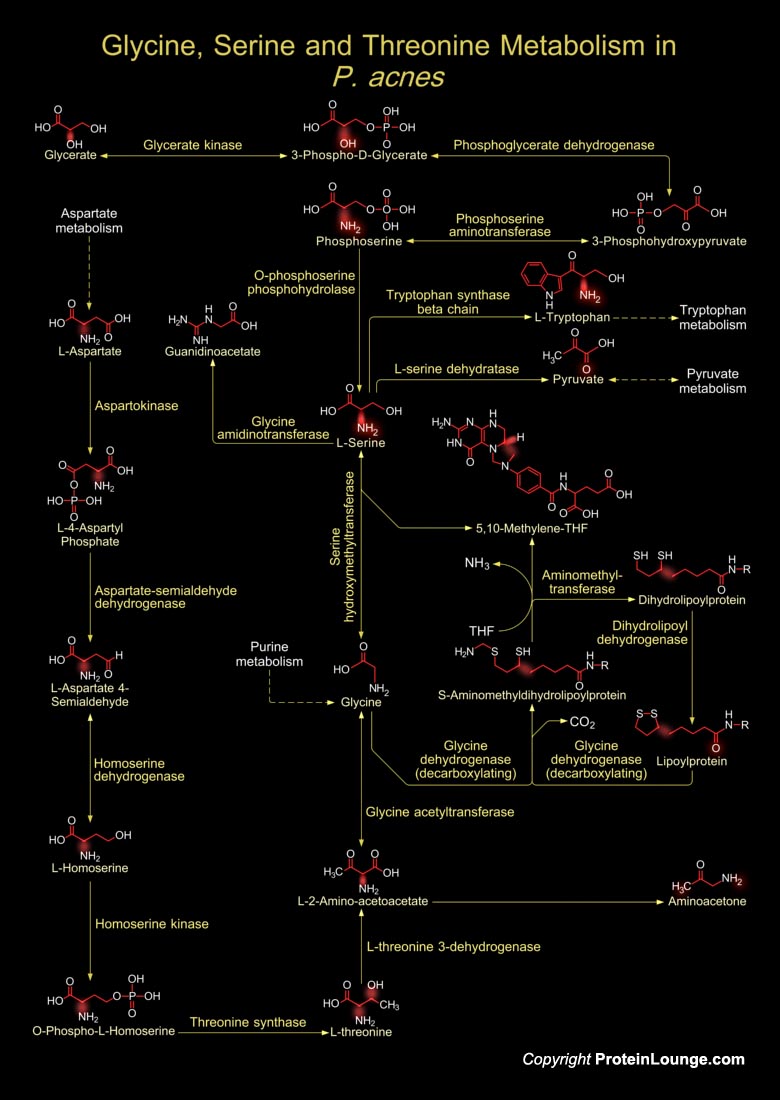
P. acnes (Propionibacterium acnes) is the most common Gram-positive, non-spore forming, anaerobic rod and a major inhabitant of adult human skin, where it resides within sebaceous follicles, usually as a harmless commensal, even though it has been implicated in Acne Vulgaris (Pimples) formation. P. acnes typically grows as an obligate anaerobe, however, some strains are aerotolerant, but still show better growth as an anaerobe. It has the ability to produce Propionic acid and Catalase along with Indole, Nitrate, or both Indole and Nitrate. The bacteria release Lipases to digest a surplus of the skin oil, Sebum. The combination of digestive products (Fatty acids) and bacterial antigens stimulates an intense local inflammation that bursts the hair follicle. Then, a[..]
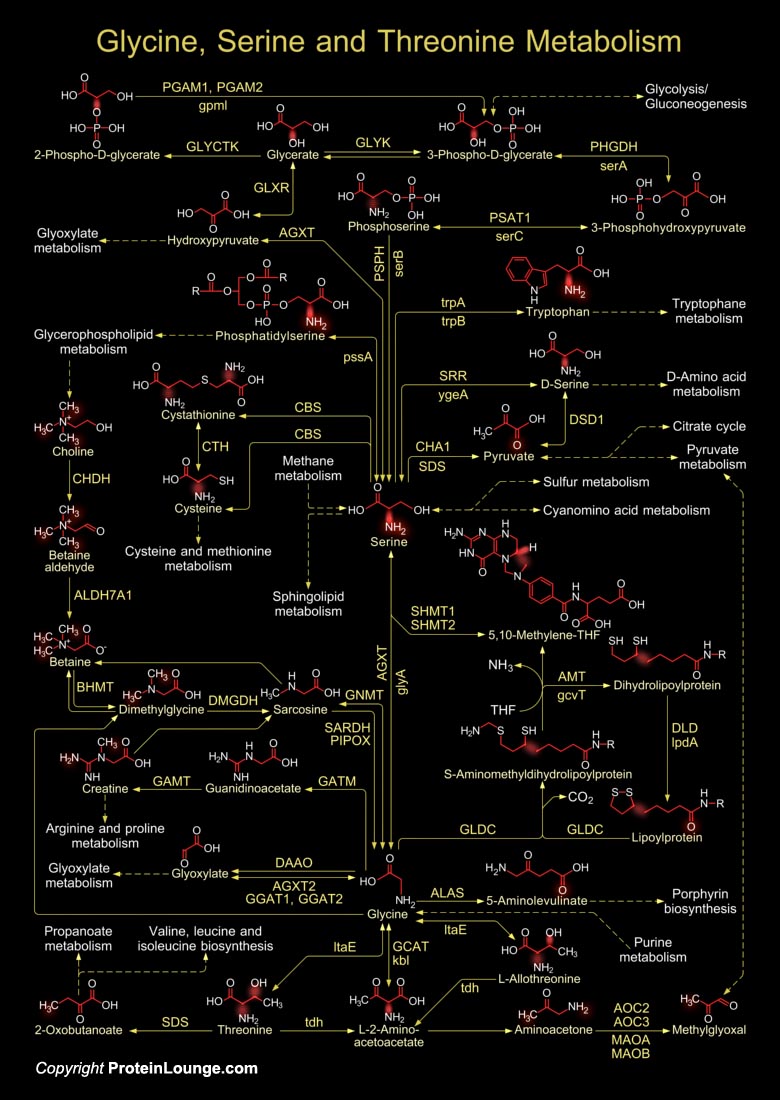
Organisms vary widely in their ability to metabolize amino acids. Based on metabolic requirements amino acids are grouped as essential amino acids (that must be provided in as nutrient) and non-essential amino acids (biosynthesized in adequate amounts). Except for Glycine, all amino acids occur in two possible optical isomers, called D and L. Because of the two hydrogen atoms at the Alpha-carbon, Glycine is not optically active. The L-amino acids represent the vast majority of amino acids found in proteins, whereas, D-amino acids are found in some proteins produced by exotic sea-dwelling organisms, but are abundant components of the cell walls of bacteria. Amino acid metabolisms are vital for the maintenance of normal nitrogen balance in an organism. The Alpha-amino[..]
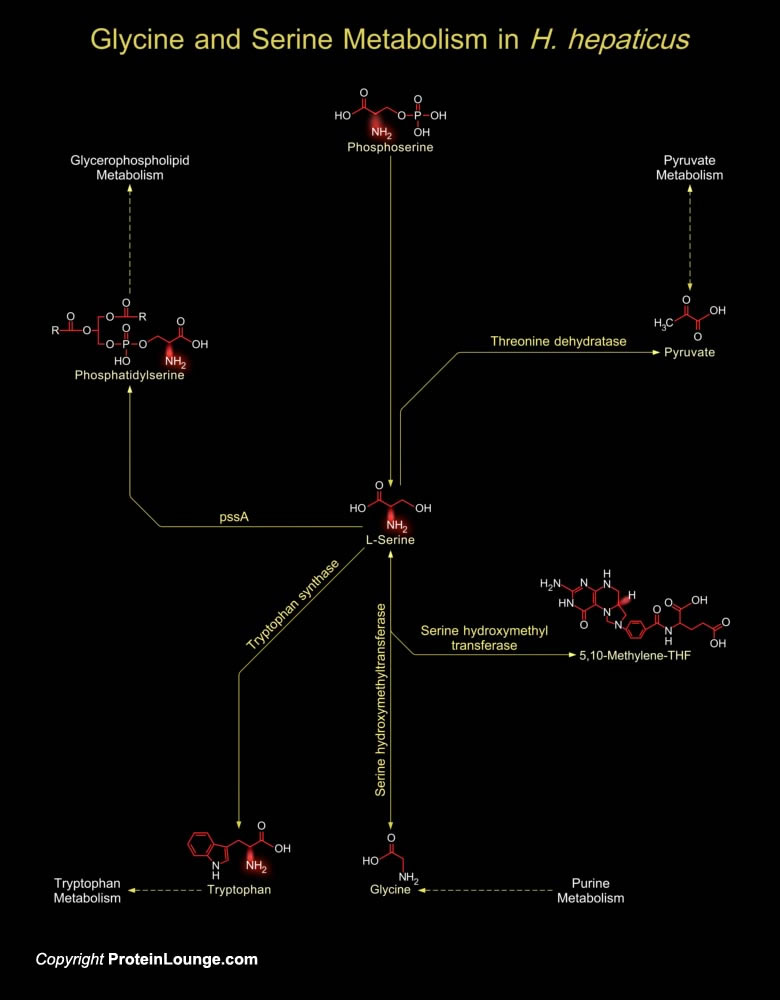
H. hepaticus (Helicobacter hepaticus) causes chronic Hepatitis and liver cancer in mice. It is the prototype enterohepatic Helicobacter species and a close relative of H. pylori (Helicobacter pylori), also a recognized carcinogen. H. hepaticus have a circular chromosome encoding 1,875 proteins. A total of 938, 953, and 821 proteins have orthologs in H. pylori, C. jejuni (Campylobacter jejuni), and both pathogens, respectively. H. hepaticus lacks orthologs of most known H. pylori virulence factors, including Adhesins, the Vacuolating Cytotoxin, and almost all cag pathogenicity island proteins, but have orthologs of the C. jejuni Adhesin Peb1 and the CDT (Cytolethal Distending Toxin) (Ref.1). The Type-IV secretion system helps in establishing pathogenicity. H. hepaticus[..]
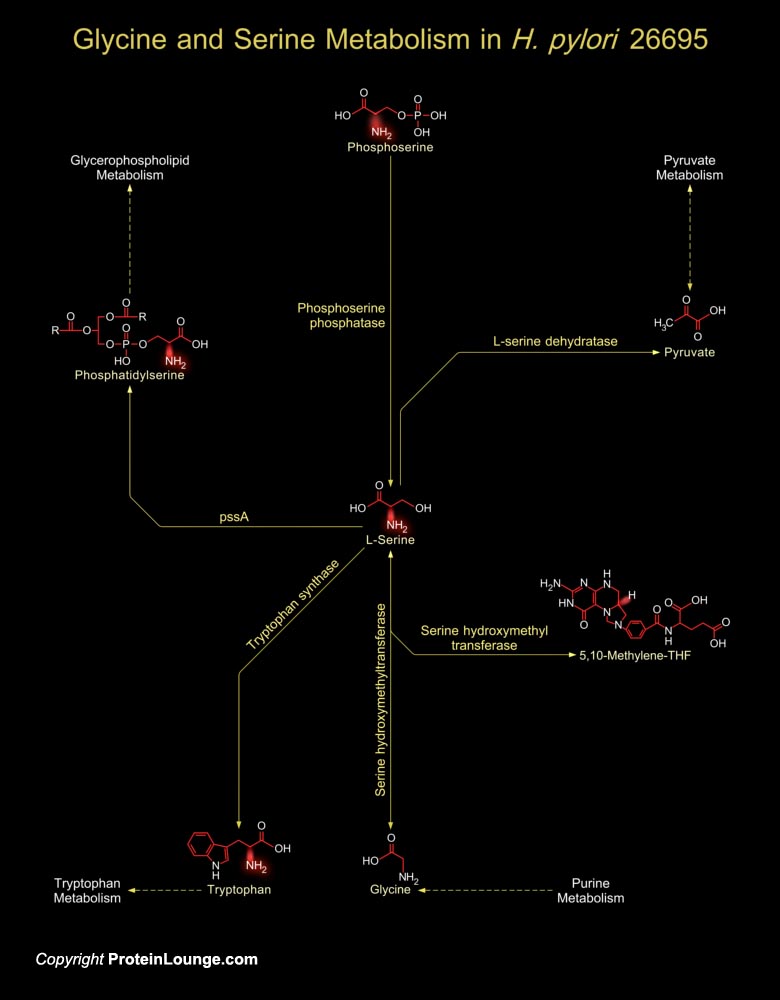
H. pylori (Helicobacter pylori) are Gram-negative, micro-aerophilic, spiral-shaped and flagellated bacteria that remains associated with Gastric inflammation and Peptic ulcer disease. As a human pathogen, H. pylori’s presence in the gastric mucosa is associated with Gastritis and is often implicated in Peptic ulceration and Mucosa-associated lymphoid tissue Lymphomas (Ref.1). The H. pylori genome is important for drug discovery and vaccine development and this is exemplified by the genome analysis of H. pylori Strain 26695. H. pylori 26695 protein-coding genes (1,590 genes) are unique to the strain. H. pylori have well-developed systems for motility, for scavenging iron, and for DNA restriction and modification. Many putative Adhesins, Lipoproteins and other[..]
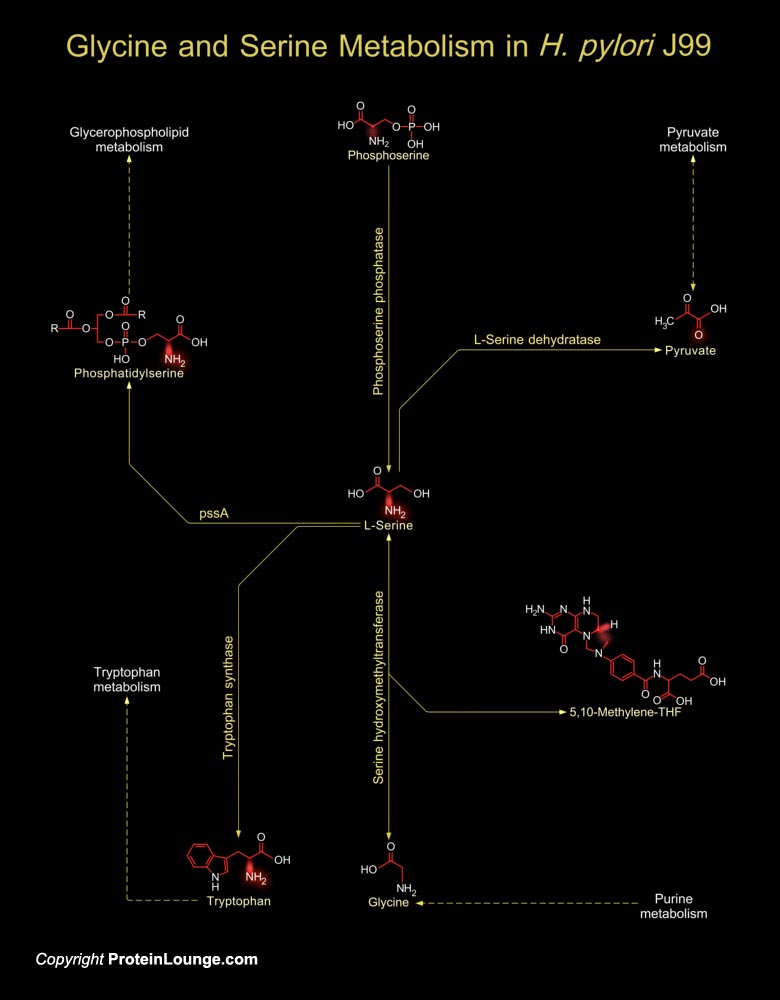
The human pathogen, H. pylori (Helicobacter pylori) present in the gastric mucosa is associated with Gastritis and is often implicated in Peptic ulceration and Mucosa-associated lymphoid tissue Lymphomas (Ref.1). The H. pylori genome is important for drug discovery and vaccine development and this is exemplified by the genome analysis of not only H. pylori Strain 26695 but also H. pylori J99. H. pylori J99 protein-coding genes (91 genes) are unique to the strain. H. pylori J99 are Gram-negative, micro-aerophilic, spiral-shaped and flagellated bacteria and mainly stimulate pathogenesis of Duodenal ulcers (Ref.2 & 3).H. pylori utilize amino acids as the sole carbon and nitrogen energy source. Amino acids like Alanine, Arginine, Asparagine, Aspartate, Glutamine,[..]
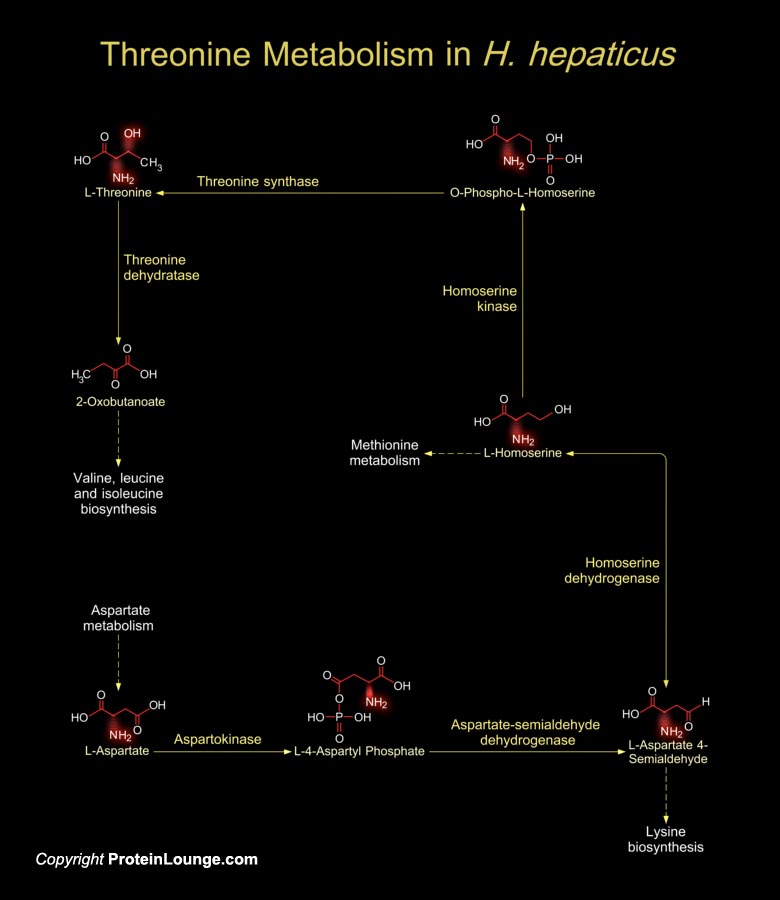
H. hepaticus (Helicobacter hepaticus) are motile and Gram-negative, curved to spiral in shape, with one to several spirals; and it has bipolar sheathed flagella (one at each end) but lacks the periplasmic fibers that envelope the bacterial cells in other mouse Helicobacter species. It grows microaerobically at 37ºC but not at 25ºC or 42ºC. H. hepaticus colonize in the large intestine and then translocate to the liver and colonize in the biliary system in mice and thereby cause chronic Hepatitis and liver cancer. It is the prototype enterohepatic Helicobacter species and a close relative of H. pylori (Helicobacter pylori), also a recognized carcinogen. H. hepaticus have a circular chromosome encoding 1,875 proteins. A total of 938, 953, and 821 proteins[..]
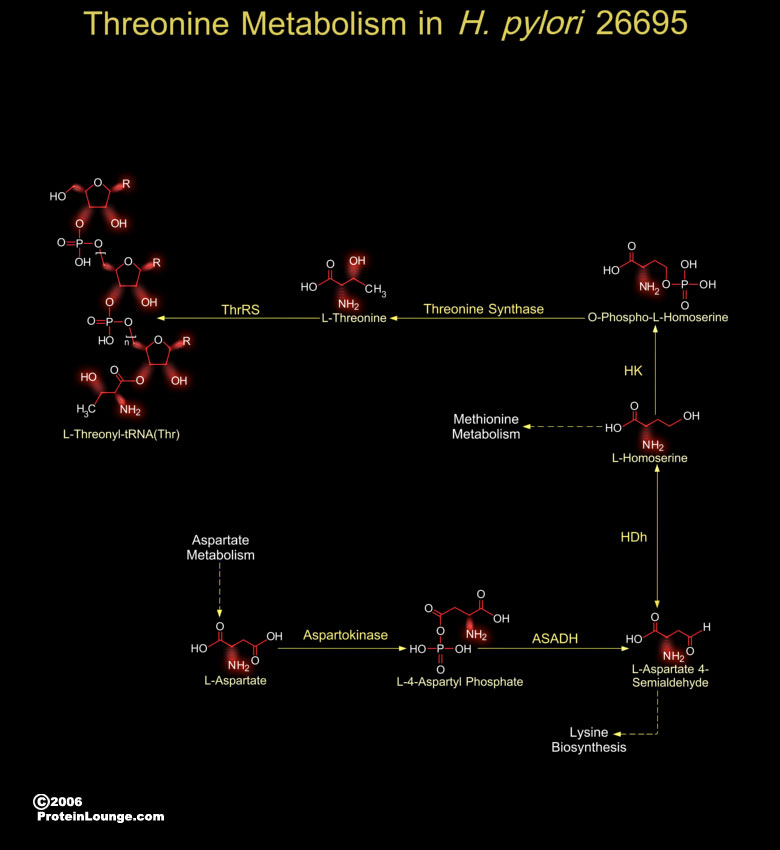
The Gram-negative, micro-aerophilic, spiral-shaped and flagellated bacteria, H. pylori (Helicobacter pylori) are associated with the pathogenesis of Gastric inflammation and Peptic ulcer disease. Presence of H. pylori in the gastric mucosa is associated with Gastritis and is often implicated in Peptic ulceration and Mucosa-associated lymphoid tissue Lymphomas. The H. pylori genome is important for drug discovery and vaccine development and this is exemplified by the genome analysis of H. pylori Strain 26695 (Ref.1). H. pylori 26695 protein-coding genes (1,590 genes) are unique to the strain. H. pylori have well-developed systems for motility, for scavenging iron, and for DNA restriction and modification. H. pylori utilize amino acids as the sole carbon and nitrogen[..]
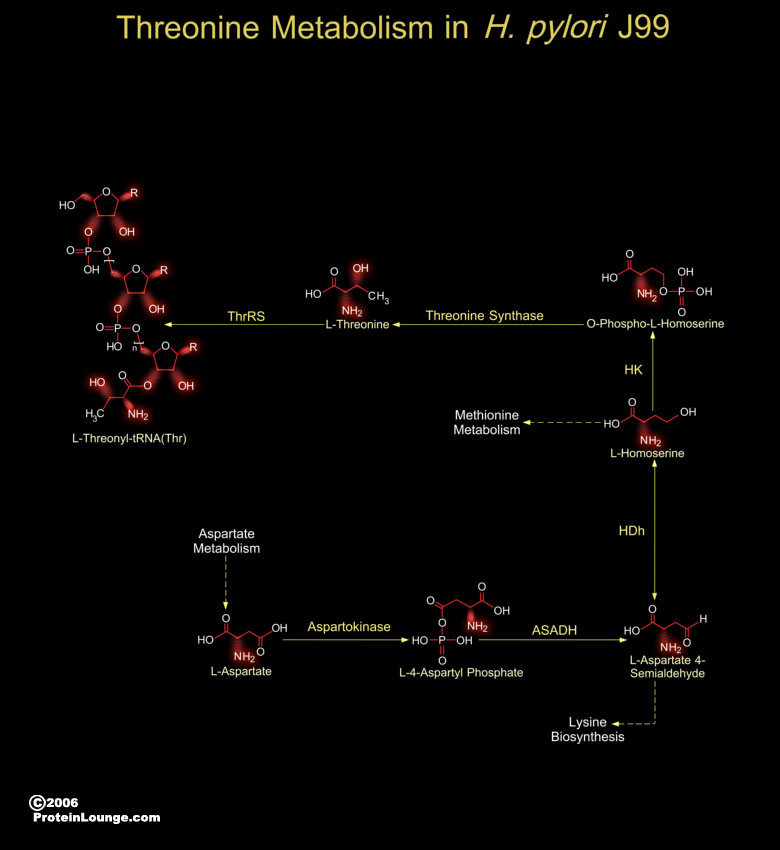
The H. pylori (Helicobacter pylori) J99 strains are Gram-negative, micro-aerophilic, spiral-shaped and flagellated bacteria, are associated with the pathogenesis of Gastric inflammation and Peptic ulcer disease (Ref.1). Presence of H. pylori J99 in the gastric mucosa is associated with Duodenal ulcers. The H. pylori genome is important for drug discovery and vaccine development and this is exemplified by the genome analysis of not only H. pylori Strain 26695 but also H. pylori J99. H. pylori J99 protein-coding genes (91 genes) are unique to the strain. H. pylori have well-developed systems for motility, for scavenging iron, and for DNA restriction and modification. H. pylori utilize amino acids as the sole carbon and nitrogen energy source. It also utilizes Glucose.[..]

