.jpg)
Diabetes mellitus is a chronic disease characterized by high blood glucose levels that requires long-term medical attention both to limit the development of its devastating complications and to manage them when they do occur. The pancreatic Beta-cell and its secretory product insulin are central in the pathophysiology of diabetes. There are two main types of diabetes, Type-I, which is insulin dependent, and Type-II, which is non-insulin dependent. Type-II diabetes is a complex and heterogeneous disease caused by both environmental and genetic factors. It maybe of various forms each of which is characterized by variable degrees of insulin resistance and Beta-cell dysfunction, and which together lead to hyperglycemia. At each end of this spectrum are single gene[..]
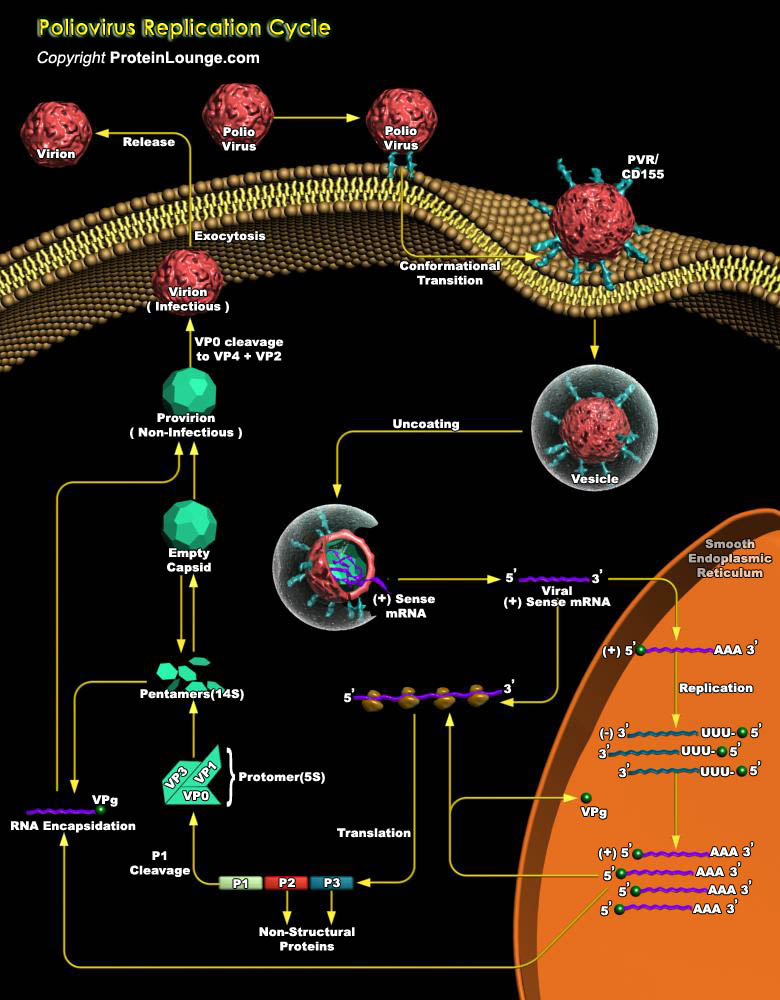
Poliovirus is a member of the Picornaviridae family, which includes a number of significant pathogens of humans (e.g., Rhinoviruses, Coxsackieviruses, Echoviruses, Enteroviruses, and Hepatitis-A virus) and livestock (e.g., foot-and-mouth disease viruses). Poliovirus has three known serotypes: PV1, PV2, and PV3 and all three serotypes can cause poliomyelitis, a paralytic disease resulting from the destruction of motor neurons in the CNS (Central Nervous System )(Ref.1). All the three serotypes of Poliovirus recognize a common cellular receptor, CD155 (or PVR, Poliovirus Receptor), for cell attachment and entry. CD155 is a trans-membrane glycoprotein belonging to the immunoglobulin super family. CD155 also functions as a cell adhesion protein and aids in transendothelial[..]
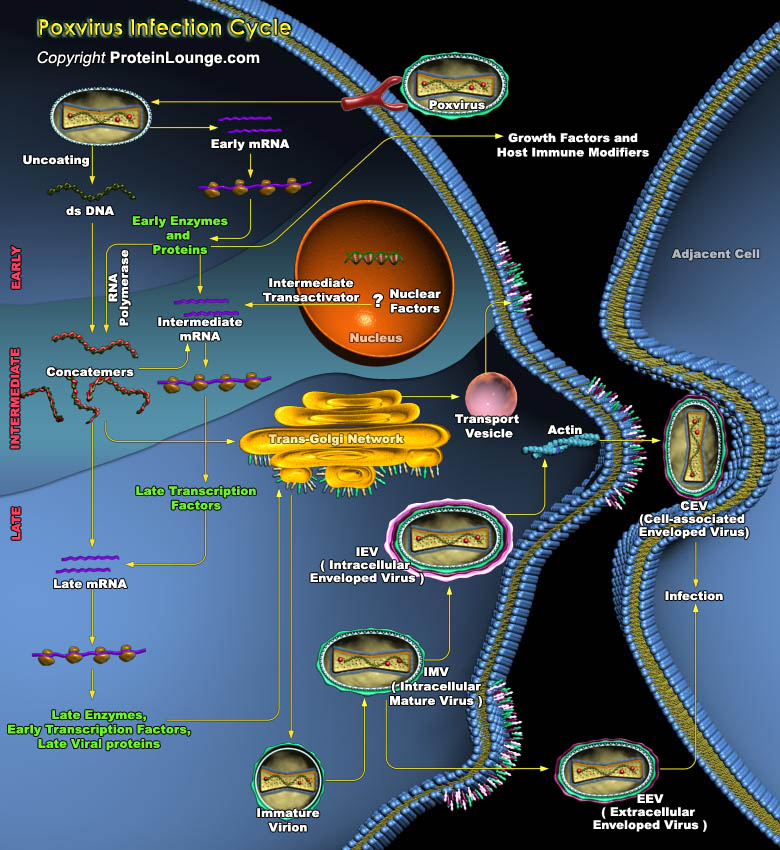
Poxviridae is a family of viruses containing large double-stranded DNA genomes of 130,000 to >300,000 nucleotides. Humans, vertebrates, and arthropods serve as natural hosts. Poxviridae viral particles (virions) are generally enveloped (external enveloped virion), though the intracellular mature virion form of the virus, which contains different envelope, is also infectious. They vary in their shape depending upon the species but are generally shaped like a brick or as an oval form similar to a rounded brick because they are wrapped by the endoplasmic reticulum. The virion is exceptionally large its size is around 200 nm in diameter and 300 nm in length and carries its genome in a single, linear, double-stranded segment of DNA. By comparison, Rhinovirus is[..]
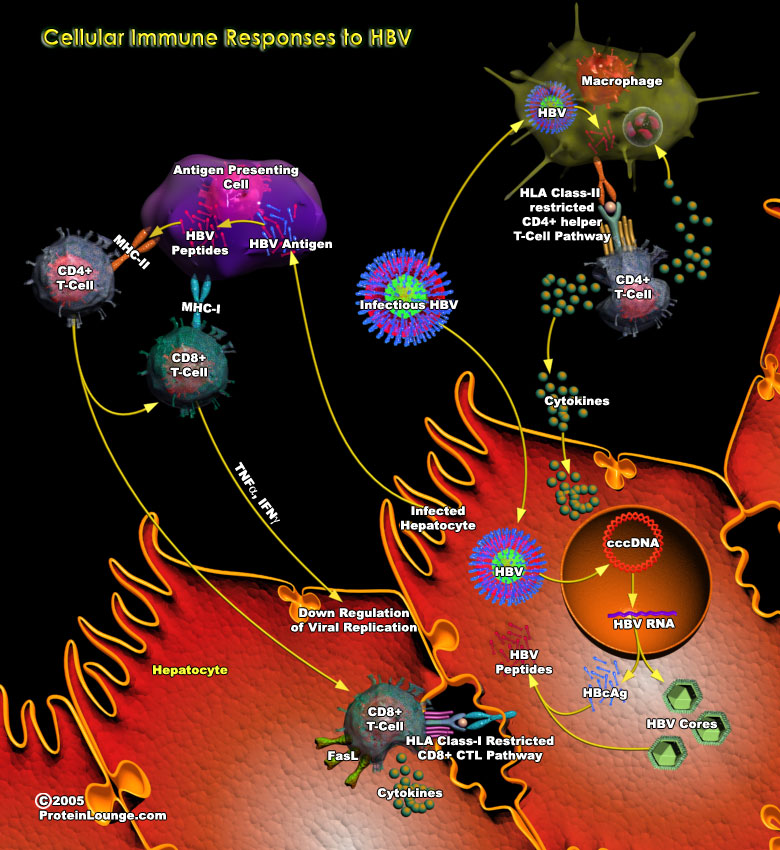
HBV(Hepatitis B virus) is a member of the family Hepadnaviridae. It is a hepatotropic non-cytopathic DNA virus and is a major cause of acute and chronic hepatitis in humans. As HBV itself is currently viewed as a non-cytopathic virus, the liver pathology associated with hepatitis B is mainly thought to be due to immune responses directed against HBV antigens. The outcome of HBV infection is the result of complex interactions between replicating HBV and the immune system.Both innate and adaptive arms of the immune system are generally involved in responding to the viral infection, with innate responses being important for control of viral replication and dissemination very early after infection, as well as for timely orchestration of virus-specific adaptive responses.[..]
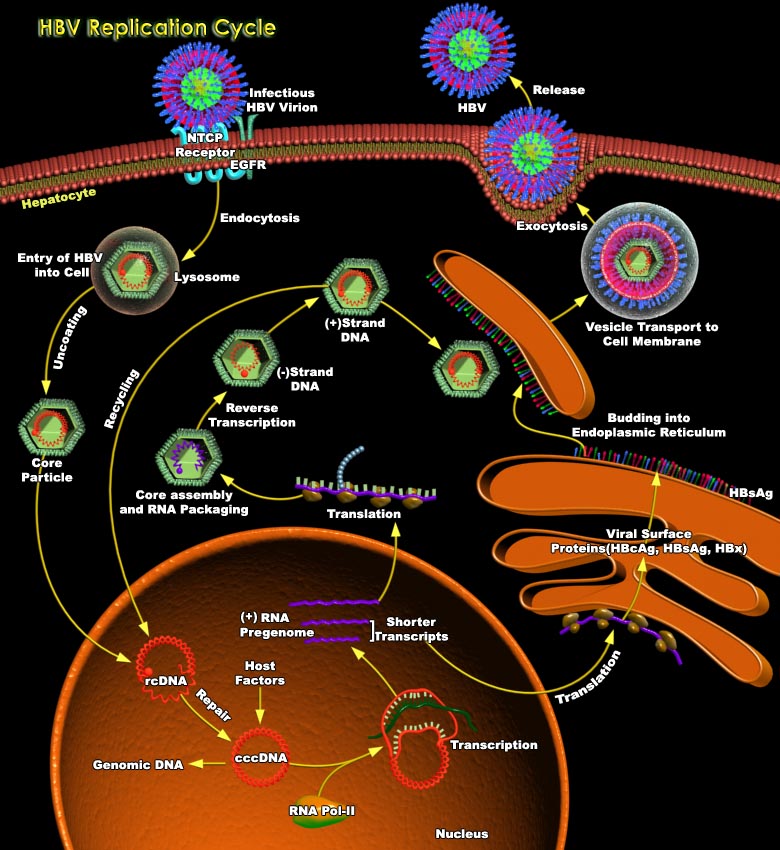
HBV (Hepatitis-B Virus) belongs to a family of closely related DNA viruses called the Hepadnaviruses. Included in this family are the WHV (Woodchuck Hepatitis Virus), the DHBV (Duck Hepatitis-B Virus) and several other avian and mammalian variants. Hepadnaviruses have a strong preference for infecting liver cells, but small amounts of hepadnaviral DNA can be found in kidney, pancreas, and mononuclear cells. However, infection at these sites is not linked to extra hepatic disease (Ref.1). The HBV nucleocapsid contains a relatively small and partially duplex 3.2 Kb circular DNA, viral polymerase and core protein. The genome has only four long open reading frames. The preS–S (presurface–surface) region of the genome encodes the three viral surface antigens by[..]
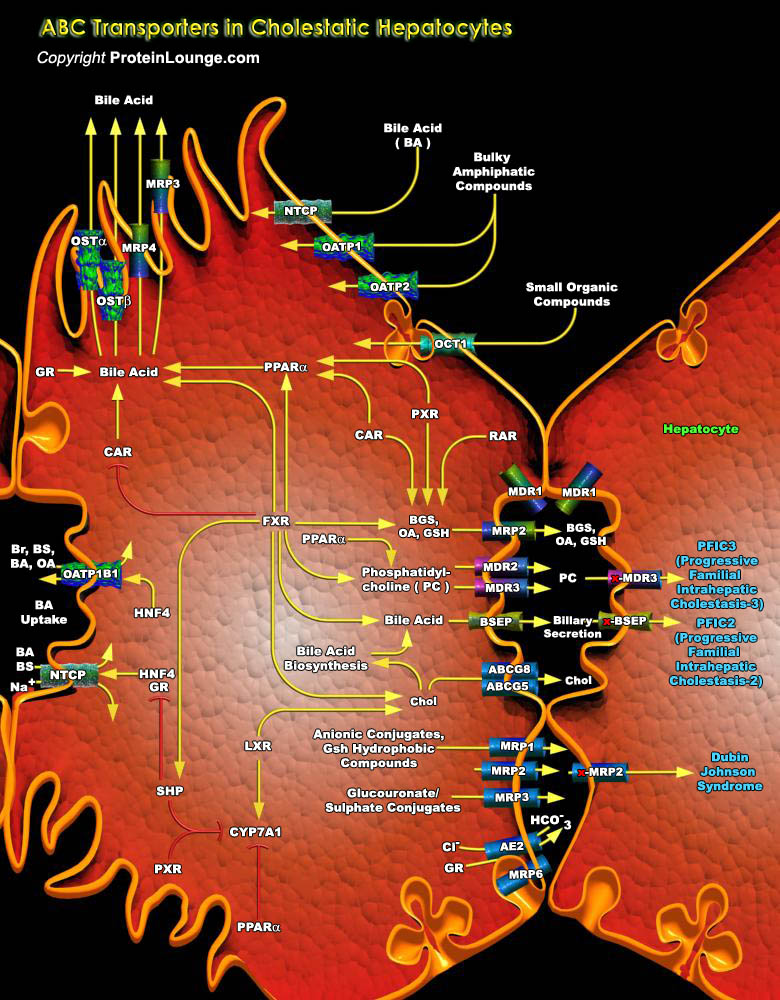
A cell must selectively translocate molecules across its plasma membrane to maintain the chemical composition of its cytoplasm distinct from that of the surrounding milieu. The most intriguing and, arguably, the most important membrane proteins for this purpose are the ABC (ATP-Binding Cassette) transporters. These proteins, found in all species, use the energy of ATP hydrolysis to translocate specific substrates across cellular membranes. The chemical nature of the substrates handled by ABC transporters is extremely diverse--from inorganic ions to sugars and large polypeptides--yet ABC transporters are highly conserved (Ref.1).Transporters have been classified as primary, secondary, or tertiary active transporters. Secondary or tertiary active transporters, such as[..]
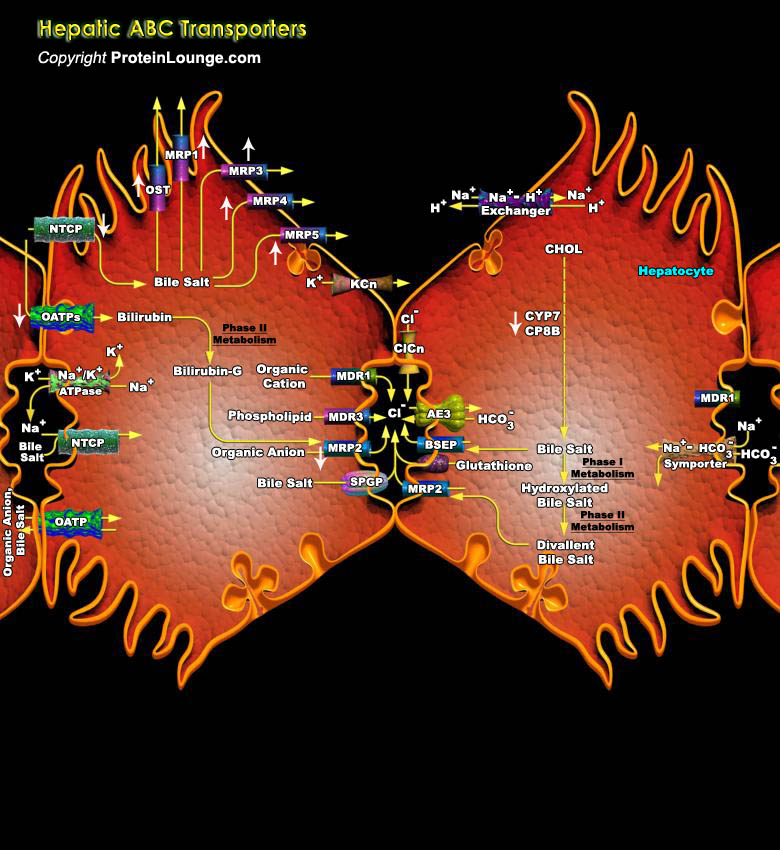
ABC (ATP-binding cassette) transporters are a large superfamily of integral membrane proteins involved in the cellular export or import of a wide variety of different substances, including ions, lipids, cyclic nucleotides, peptides, and proteins. ABC transporters are systemically classified into eight subfamilies by sequence similarity, i.e., ABCA (ABC1), ABCB (MDR/TAP), ABCC (MRP/CFTR), ABCD (ALD), ABCE (RNAseLI/OABP), ABCF (GCN20), ABCG (White) and ANSA subclass. In general, the transmembrane part of ABC transporters contains a polar channel formed by two homologous domains, each usually consisting of five (uptake transporters) or six (efflux transporters) transmembrane alpha-helices (Ref.1 & 2). Typically, ABC proteins are relatively specific for a[..]
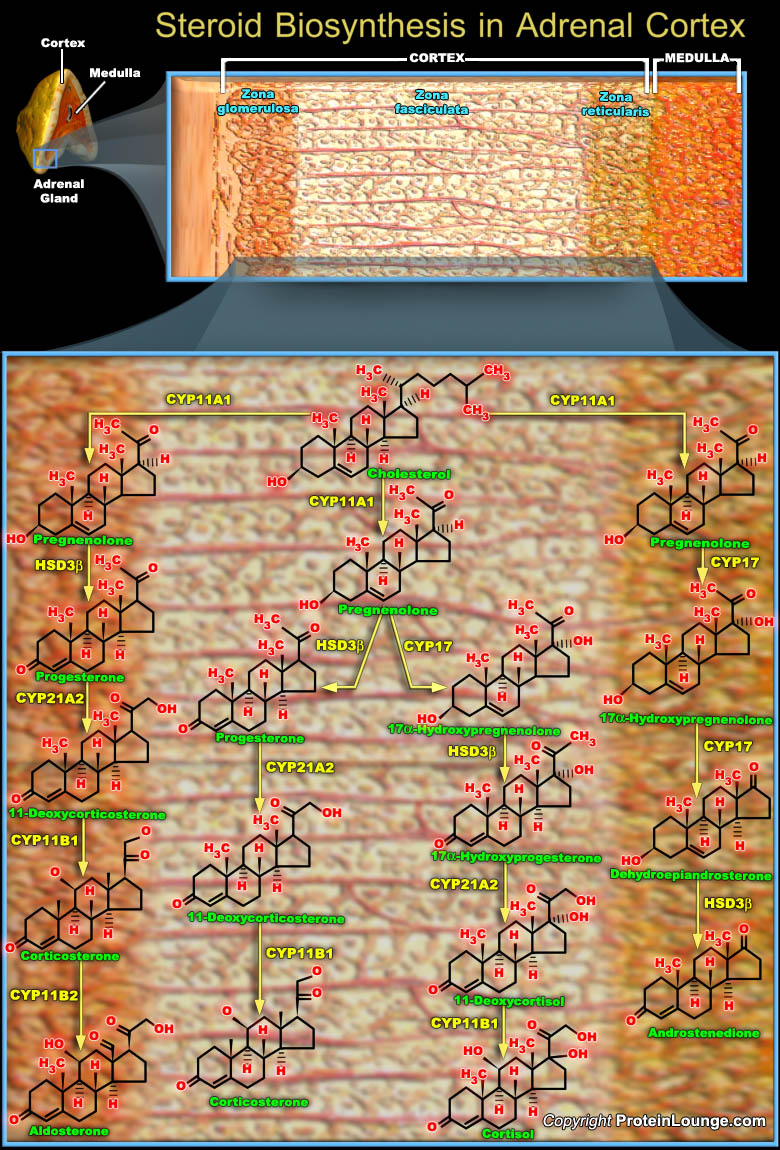
Steroid hormones are lipophilic, low-molecular weight compounds derived from Cholesterol that play important physiological roles. The steroid hormones are synthesized mainly by Endocrine Glands such as the the Adrenal Cortex and the Gonads (Ovary and Testes), and are then released into the blood circulation. There are five major classes of steroid hormones. They are the (i) Glucocorticoids (Anti-Stress Hormones), Cortisol is the major representative in most mammals; Mineralocorticoids (Na+ Uptake Regulators), Aldosterone being most prominent; Androgens (Male Sex Hormones), such as Testosterone; Estrogens (Female Sex Hormones), including Estrodiol and Estrone; and, Progestogens (Progestational hormones), such as Progesterone. The Adrenal Cortex is responsible for[..]
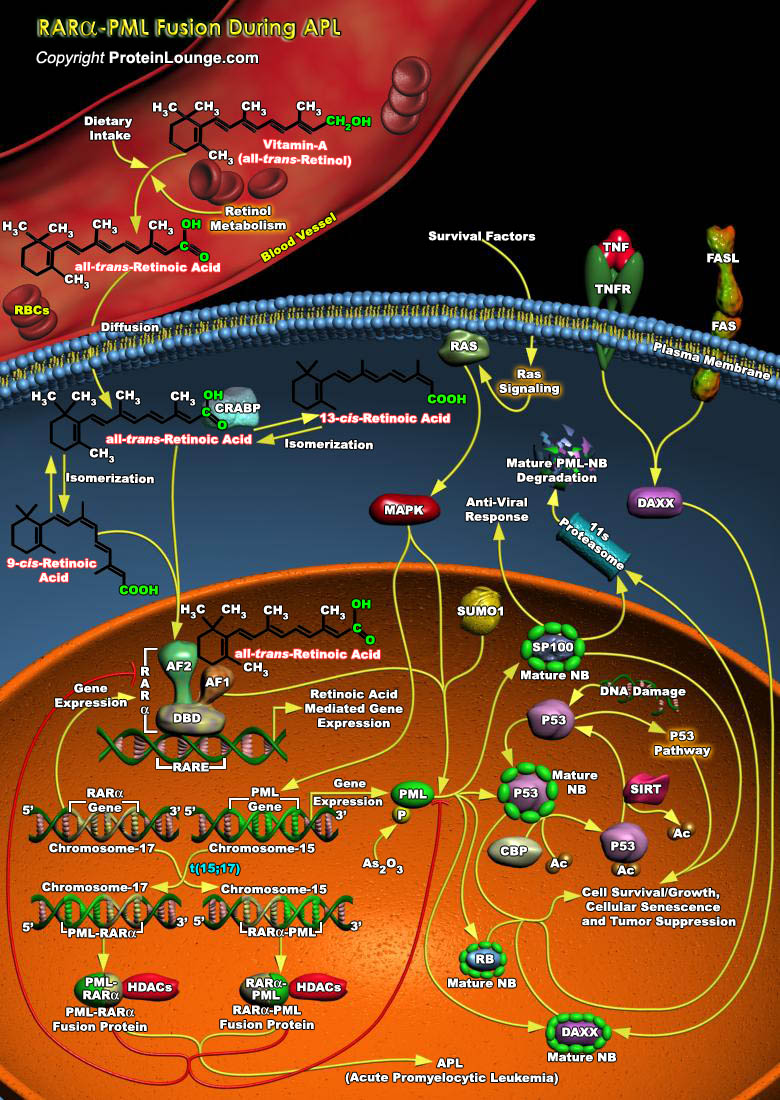
AMLs (Acute Myeloid Leukemias) are characterized with chromosomal translocations resulting in the formation of fusion proteins. Understanding PML (Acute Promyelocytic Leukemia Inducer) function has become an area of intense research because of its involvement in the pathogenesis of APL (Acute Promyelocytic Leukemia), a distinct subtype of Myeloid Leukemia. In the vast majority of APL case studies, the PML gene (on Chromosome-15) fuses to the RAR-Alpha gene (Retinoic Acid Receptor-Alpha) (on Chromosome-17) as a consequence of reciprocal and balanced chromosomal translocations. In the t(15;17) chromosomal translocation, which is specific for APL, PML is found in a reciprocal translocation with the RAR-Alpha resulting in the formation of PML-RAR-Alpha and RAR-Alpha-PML[..]
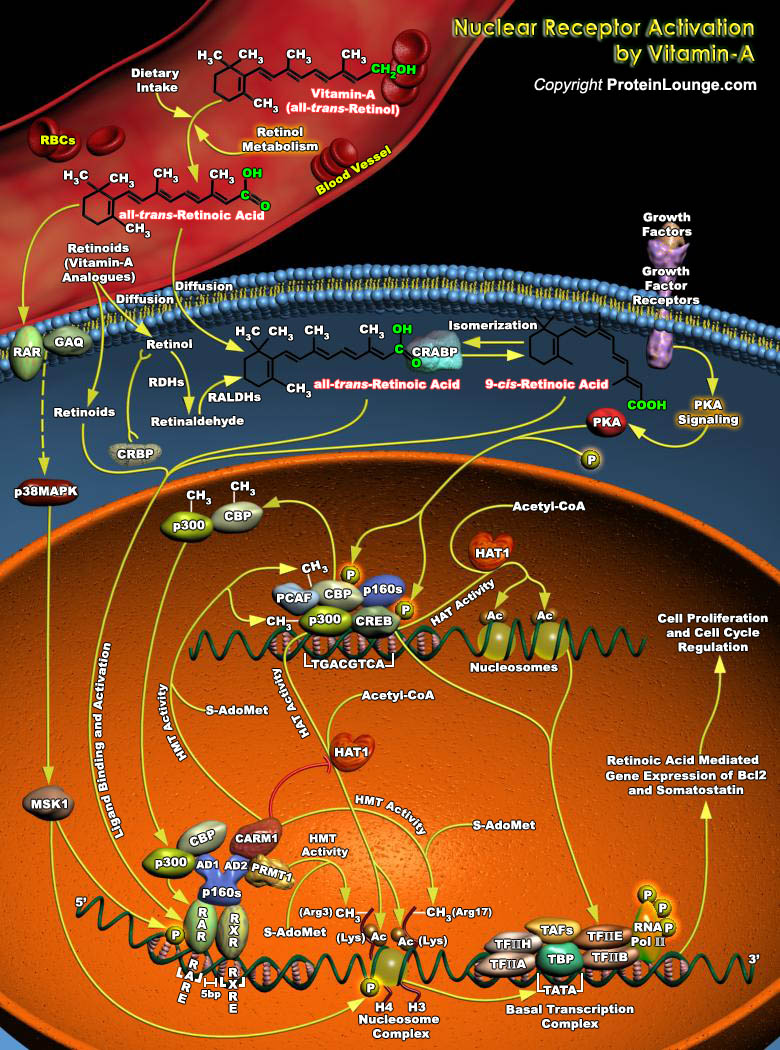
Vitamin A and its analogs, collectively termed retinoids, have a profound effect on cell growth, differentiation, apoptosis, and morphogenesis. Retinol, the lipid-soluble vitamin A, is an absolute requirement for normal growth, vision and differentiation of epithelial tissues in mammals. Retinol must be obtained directly through dietary intake, but may also be derived in its provitamin A forms obtained through dietary carotenoids (Ref.1 and 2). Retinoids bind to six distinct nuclear receptors in mammals and regulate the expression of target genes. The Retinoic Acid Receptors (RARs) and Retinoid X Receptors are among the most intensely studied nuclear hormone receptors. RARs are ligand-controlled transcription factors that function as heterodimers with RXRs to regulate[..]
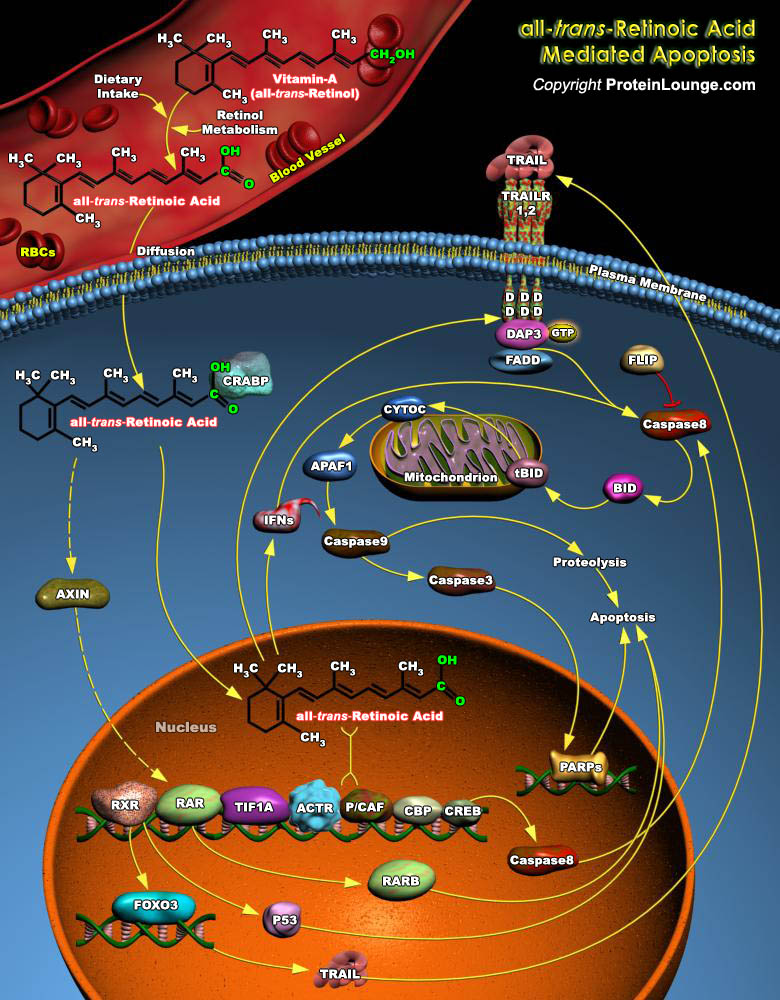
Retinoic Acid, a lipophilic molecule and a metabolite of Vitamin-A (all-trans-Retinol), affects gene transcription and modulates a wide variety of biological processes like Cell Proliferation, Differentiation, including Apoptosis. Retinoic Acid mediated gene transcription depends on the rate of transport of Retinoic Acid to target cells and the timing of exposure of Retinoic Acid to RARs (Retinoic Acid Receptors) in the target tissues. The all-trans-Retinoic Acid, the Carboxylic Acid form of Vitamin-A is of biological significance since it has high circulating levels than other isomers of Retinoic Acid. The targets of all-trans-Retinoic Acid and RARs include a multitude of Structural genes, Oncogenes, Transcription Factors and Cytokines. Although biologically active[..]
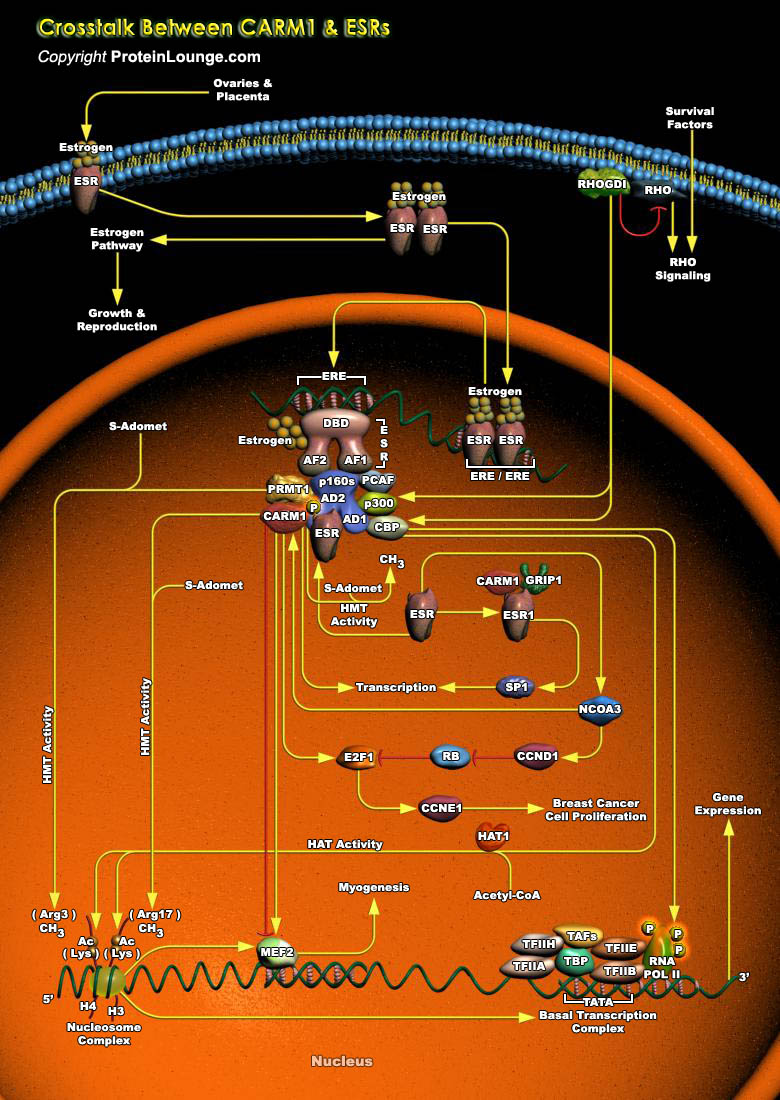
The ESRs (Estrogen Receptors) are ligand-dependent transcription factors and are important Nuclear Hormone Receptors that act as regulators of cell growth, differentiation and malignant transformation. Transcriptional activation by ESRs is accomplished through specific and general cofactor complexes that assemble with the receptor at target promoters to regulate transcription. The chief ligand for ESR is the ovarian steroid hormone Estrogen, which has a primary role in the establishment and maintenance of reproductive function (Ref.1). Naturally occurring forms of Estrogen are Estradiol, Estriol, and Estrone. Estradiol is the most commonly occurring form of Estrogen in non-pregnant women. Binding of Estrogen to the ESR promotes a conformational change in the receptor[..]

